Identification of Patterns in Coronavirus Vaccines: Evidence of DNA-Origami Self-Assembly
Identification of Patterns in Coronavirus Vaccines: Evidence of DNA-Origami Self-Assembly
by Mik Andersen, C0r0n@2Inspect
published in Spanish January 3, 2022
rough translation via translation software
One of the most difficult aspects to determine in the identification of patterns of c0r0n@v|rus vaccines is the method or procedure, by which the objects that are being observed ( micro / nano-routers, micro / nano-rectenas . ..), have been able to conform. In the scientific literature, a multitude of works have been found that pointed to various production techniques, such as electron lithography, focused ion beam FIB (Focused Ion Beam) and even synthetic DNA templates, with which QCA circuits would be defined. of the nanorouters.
However, no clear evidence of self-assembly was found in the vaccine samples. However, the suspicions, more than founded on this process, were confirmed with the observation of the video produced by Ricardo Delgado on December 27, 2021 , in which the movement of thousands of particles in a sample of the Pfizer vaccine was witnessed. These particles seemed to come together to form more complex structures, defining simple geometric patterns, see excerpt in the following video 1.
https://www.twitch.tv/videos/1245191848?t=00h34m56s (Delgado, R. 2021)
In the scientific literature, this quasi-directed behavior or movement of the particles, in the context of the construction of micro / nano electronic objects and devices in an intracorporeal communications nano-network, had a high probability of corresponding to a self-assembly process based on DNA, epitaxial growth and origami. This deduction resulted in the location of the scientific article that, with high probability, could confirm the self-assembly of complex objects, including circuits, boards, routers, sensors and other micro / nano electronic components and devices.
This discovery explains how the components responsible for the bluetooth MAC address emission phenomenon would self-assemble (Sarlangue, G.; Devilleger, J.; Trillaud, P.; Fouchet, S.; Taillasson, L.; Catteau, G. 2021) .It would also explain the assembly of nano-devices,nano-sensors, nano-nodes, micro / nano-interfaces,micro / nano-routers , micro / nano-antennas , micro / nano-rectenas , with which configures the network hardware intra-body of nano-communications .
Figure 1 shows the signs of self-assembly observed in the scientific literature and its correspondence with the analyzed samples of the Pfizer vaccine. From a morphological point of view, there are important coincidences that allow us to infer and almost assume that self-assembly is a verifiable reality.
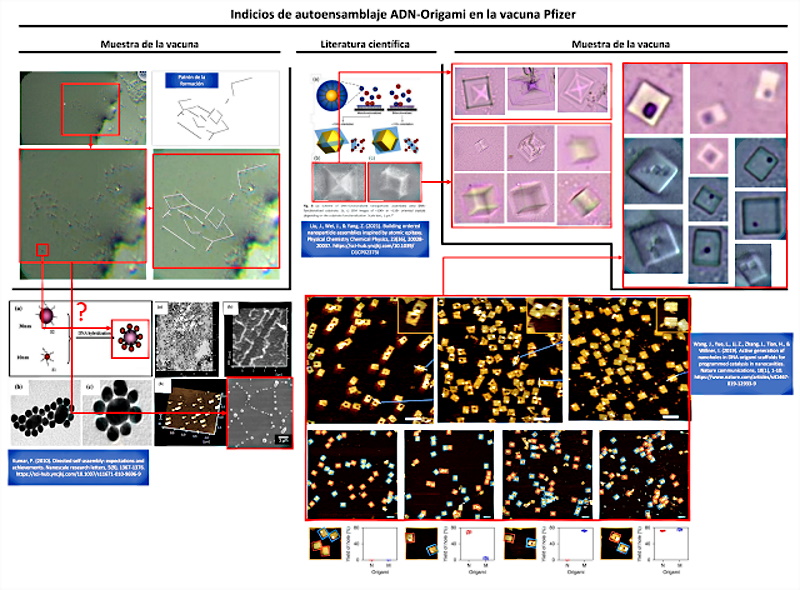
Due to the complexity of the self-assembly issue, as well as the relevance of the evidence discovered, a detailed analysis will be carried out, around three fundamental headings: a) directed self-assembly; b) self-assembly by smooth epitaxial growth; c) origami self-assembly.
Directed self-assembly
The article by (Kumar, P. 2010) presents the first clear indication of “directed self-assembly” that can be observed in the vaccine sample, see figure 2 and video 1. The observed nanoparticles seem to unite in clusters of larger size and with it, more complex structures that move in the sample drop.

According to (Kumar, P. 2010), the self – assembly directed is key to the development of electronic devices, magnetic and optical miniaturized, which fits with materials derived from graphene found in samples of the vaccine, in fact states that “the Nanoparticles have attracted a great deal of attention as such components due to their unique size-dependent properties, including super-paramagnetism, chemiluminescence, and catalysis. To take full advantage of the potential capabilities of nanoparticles, we need to develop new methods to assemble them into useful patterns or structures. These self-assembling structures promise new opportunities to develop miniaturized optical, electronic, optoelectronic and magnetic devices .”
On the other hand, Kumar reveals that the “directed self-assembly” method is suitable for generating nano- and micro-scale devices due to its ability to use quantum dots or nanopoints. He explains it in the following way ” as the size or functions of the device get smaller and smaller, conventional lithographic processes turn out to be limited for their production. It is necessary to develop alternative methods to overcome this difficulty. As they are developed Conventional manufacturing technologies, such as optical lithography, are also beginning to encounter fundamental limits … In addition, new manufacturing techniques are required to help extend both the life and range of application of existing techniques … Directed self-assembly technique can be used appropriately to produce functional nanostructures, for example nanowires and an organized array of nano-dots (understand quantum dots) . “In other words,” directed self-assembly “allows quantum dots to of a certain material (for example the graphene GQD Graphene Quantum Dots), they self-assemble according to a predefined pattern.
Among the possible types of directed or guided self-assembly, Kumar recognizes the “assembly guided by templates where they use atomic surface patterns; the assembly guided by electromagnetic field or electric field, by electron beam, light and laser, among others“.
Furthermore, it recognizes that “directed self-assembly is a robust and reproducible technique with future prospects for use on an industrial scale … which means building well-ordered, often intriguing structures, which has received a lot of attention for its ease of organizing materials. at the nanoscale in ordered structures and produce complex structures on a large scale.” This seems to be fundamental in the context of the intra-corporal network of nanocommunications and nano / micro devices, since thousands of devices must be created for their operation ( Zhang, R.; Yang, K.; Abbasi, QH; Qaraqe, KA; Alomainy, A. 2017 | Galal, A.; Hesselbach, X. 2018 | Galal, A.; Hesselbach, X. 2020)
Among all the forms of self-assembly, the most probable and the one with the greatest coincidences at the morphological level is self-assembly guided by biological DNA templates. Among its advantages, Kumar highlights “the manufacture of nanowires since they solve integration problems (eliminating the need to manipulate individual nanowires). They also solve problems related to contacts for electrical and magnetic transport.” This fits with the type of nanodevices observed, for example micro / nano rectenas and graphene-derived materials, GQD graphene quantum dots.
In fact, Kumar states that “the use of physical DNA templates, results in the growth of nanomaterials in a predefined position, eliminating the need for post-growth manipulation and providing the ease of electrical connections for additional characterizations“, which helps to understand how the structures are constructed and defined. Quadrangular shapes observed in the vaccine samples, which bear a great resemblance to PCBs, microchips, sensors and integrated circuits. He also adds that ” such templates give rise to the growth of nanopoints (quantum dots), vertical nanowires, which can be used in a controllable way to manufacture FET devices (Field Effect Transistors), magnetic tunnel junction devices and devices for optical applications” which confirms that with directed self-assembly it is possible to create miniaturized nanotechnology of any known electronic device.
In other words, self-assembly guided by biological DNA templates can be used to make all the devices required for an intracorporeal nano-network , being feasible that this is the technique used in vaccines, according to the observed images and the statements in the scientific literature (Catania, V .; Mineo, A .; Monteleone, S .; Patti, D. 2014 | Keren, K .; Berman, RS; Buchstab, E .; Sivan, U .; Braun, E. 2003).
To be exact, Kumar indicates that “l as directed strategies biomolecules (biological DNA templates) have shown great promise in the nanoparticle assembly in a wide variety of architectures, because of its high efficiency, high specificity and genetic programmability ( McMillan , RA; Paavola, CD; Howard, J .; Chan, SL; Zaluzec, NJ; Trent, JD 2002 ). These nanoassembled materials have been shown to have potential applications in new detection systems, such as biosensors ( Taton, TA; Mirkin , CA; Letsinger, RL 2000 ) and chemical sensors ( Liu, J .; Lu, Y. 2003 | Liu, J .; Lu, Y. 2006 ), and in the construction of nanoelectronic devices ( Keren, K .; Berman, RS; Buchstab, E .; Sivan, U .; Braun, E. 2003 ) [paradoxically configured with carbon nanotubes] ” Which again confirms that it is a convenient technique / method in the implementation of nanotechnology in the human body.
Self-assembly through smooth epitaxial growth
If evidence of directed self-assembly can be considered a well-founded hypothesis, “self-assembly by smooth epitaxial growth” presents even more compelling evidence. Figure 4 shows an exact equivalence between the scientific literature and the Pfizer vaccine samples analyzed by the doctor (Campra, P. 2021). Some of the most numerous objects, with a quadrangular and pyramidal shape, would actually be the result of an epitaxial self-assembly technique, which is in fact, “one of the manufacturing processes of integrated circuits” (Shen, J .; Sun, W .; Liu, D .; Schaus, T .; Yin, P. 2021 | Burns, MA; Mastrangelo, CH; Sammarco, TS; Man, FP; Webster, JR; Johnsons, BN; Burke, DT 1996 | Esener, SC; Hartmann, DM; Heller, MJ; Cable, JM 1998 | Krahne, R .; Yacoby, A .; Shtrikman, H .; Bar-Joseph, I .; Dadosh, T. ; Sperling, J. 2002 | Chen, Y .; Pepin, A. 2001 ) Epitaxy refers to the deposition of a layer of material (for example quantum dots of graphene, graphene oxide, hydrogel, etc.) on a Primary nucleation substrate. However, unlike traditional growth processes, in this case it is achieved through DNA hybridization. It is at this point, where (Liu, J .; Wei, J .; Yang, Z. 2021 ) develops one of the objects of his research.
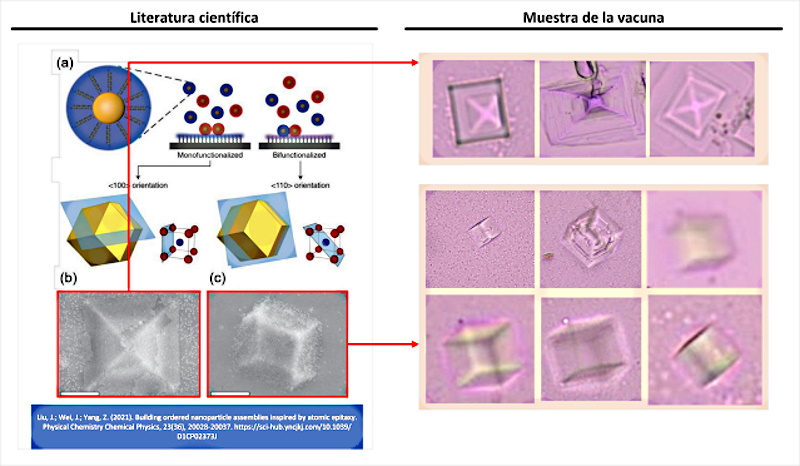
According to (Liu, J.; Wei, J.; Yang, Z. 2021) the self-assembly of “inorganic nanoparticles into mesoscopic or macroscopic nanoparticle assemblies is an efficient strategy to manufacture advanced devices with emerging nanoscale functionalities. In addition, the assembly of Nanoparticles on substrates can allow the fabrication of substrate-integrated devices, similar to the growth of atomic crystals on a substrate. Recent progress in nanoparticle assembly suggests that ordered nanoparticle assemblies could well be produced on a selected substrate, which known as smooth epitaxial growth.”
This definition confirms that the manufacture of micro / nano electronic devices (integrated circuits) can be carried out by guided crystal growth on a DNA substrate or template. This is evident in the following explanation “DNA hybridization It has been applied to assemble nanoparticles into superlattices with crystalline structures that are surprisingly rich. The three-dimensional double helix structure of DNA (fixed pitch, fixed diameter) was found to have more advantages than other materials in guiding nanoparticles toward an ordered three-dimensional assembly (Nykypanchuk, D.; Maye, MM; Van-Der-Lelie , D .; Gang, O. 2008). Specific recognition between base pairs and the ability to control DNA strand length and base sequence make it a powerful weapon for nanoscale assembly. The programming capacity of DNA makes it an extremely attractive structure-oriented ligand .” This confirms that self-assembly by means of DNA not only allows the construction of 2D structures, since 3D structures can be generated thanks to the bonds of the double helix of the DNA, which allows it to be used to configure all kinds of shapes , including cubic and prismatic shapes seen in figure 4.
Among the experiences cited by (Liu, J .; Wei, J .; Yang, Z. 2021) it is worth highlighting the following paragraph on epitexial self-assembly, in which a wide experience in the experimentation of DNA- based crystalline constructions is revealed, with a tolerance to error (mismatch) of only 1%.
“According ( Lewis, DJ; Zornberg, LZ; Carter, DJ; Macfarlane, RJ 2020 ) and colleagues used this technique and a combination of nanoparticles functionalized with DNA and a substrate functionalized DNA strand to design a process of epitaxial assembly . They found that monocrystalline Winterbottom forms of nanoparticle crystals are formed by controlling the interfacial energies between crystals and fluid, substrate and crystal, and substrate and fluid . Other examples show that grafted DNA nanoparticles self-assemble into two-dimensional colloidal films can be applied as a substrate for smooth epitaxial assembly . For example, according to ( Wang, MX; Seo, SE; Gabrys, PA; Fleischman, D .; Lee, B .; Kim, Y .; Mirkin, CA 2017 ) used DNA-coated nanoparticles as more elastic and malleable building elements to better adapt to network mismatch . Later studies ( Gabrys, PA; Seo, SE; Wang, MX; Oh, E.; Macfarlane, RJ; Mirkin, CA 2018) showed that superlattice thin films assembled by DNA functionalized nanoparticles can store elastic strains by deforming and reorganizing, with lattice mismatches of up to ± 7.7%, significantly overcoming the ± 1% lattice mismatches allowed by atomic thin films.It is important to highlight that these DNA-coated nanoparticles experience a progressive and coherent relaxation, dissipating the tension in an elastic and irrecoverable way through the formation of dislocations or vacancies. Therefore, it is possible to grow heteroepitaxial colloidal films by controlling programmable -soft- atomic equivalents of nanometers and microstructures using rigid nanocrystals coated with soft compressible polymeric materials.” (Liu, J.; Wei, J.; Yang, Z. 2021)
Self-assembly origami
Finally, among the most original forms of self-assembly is the “origami method“, also linked to the use of DNA templates. In this case, the evidence is found in the work of ( Wang, J.; Yue, L.; Li, Z.; Zhang, J.; Tian, H.; Willner, I. 2019 ) entitled “Active generation of nano -holes in DNA origami scaffolds for programmed catalysis in nanocavities“. The pattern of a point or hole within a quadrangular structure is striking and characterizing from the morphological point of view. This detail was found in the images obtained by Dr. Campra, which together with the self-assembly study object, allowed infer that it was another piece of the puzzle and that in reality, there must be larger objects self-assembled with the origami method. The similarities are clear and evident, see figure 5, since the quadrangular structure of the objects coincides, the position of the nano -holes inscribed within the surface, as well as the number or quantity of them observed in the Pfizer vaccine samples.
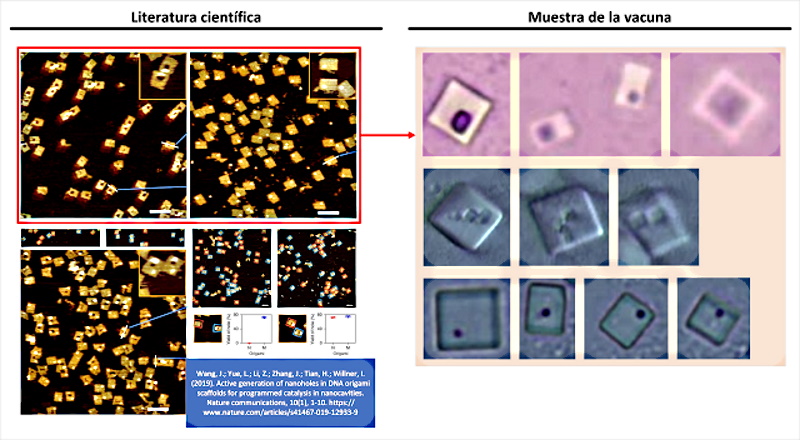
But, before proceeding to analyze the issue of holes in quadrangular objects, it is worth reviewing the introduction and state of the art provided by the authors in their work, as it helps to locate the capabilities of the technique and demonstrate its link to nanotechnology used in vaccines. In fact, surprising claims are made, since origami self-assembly is a “programmed assembly of two-dimensional (2D) and three-dimensional (3D) DNA nanostructures, representing a major advance in DNA nanotechnology” (Hong, F .; Zhang , F .; Liu, Y .; Yan, H. 2017 | Rothemund, PW 2006 | Endo, M .; Sugiyama, H. 2014), which confirms not only the possible dimensions or axes of self-assembly, but also that the origami method is compatible with self-assembly with smooth epitexial growth and therefore with directed or guided self-assembly. In all cases, the use of suitably configured synthetic DNA structures are the necessary precursors for the development of the structures and objects observed in the vaccine samples.
In addition, (Wang, J .; Yue, L .; Li, Z .; Zhang, J .; Tian, H .; Willner, I. 2019) confirm that the origami self-assembly method using DNA allows the anchoring of components for configure, among other devices, plasmonic antennas , previously identified in the vaccine samples as part of the nano-network centered on the human body . This is stated in the following literal quote: “In addition to creating ingenious forms of origami structures generated by the programmed folding of DNA, origami structures were functionalized with protruding nucleic acid strands, or strands of oligonucleotides with modified edges. The protruding strands were used as anchor sites for the organization of the polymers, proteins and nanoparticles in the scaffolds of each origami. Unique functions of assembled nanostructures in origami scaffolds were demonstrated, such as the operation of enzyme cascades, the design of plasmonic antennas, and the assembly of chiroplasmonic structures.”
This explanation is essential to understand the process of formation of superstructures, guided by DNA patterns, since they are linked through the strands that protrude from the building pieces, functionalized with nanoparticles (for example graphene quantum dots), which together with the scale factor and superconductor of the material, provide plasmonic characteristics, and quantum hall, which implies the self-assembly of transistors, and micro / nano chips of the complexity that is required.
In their introduction, (Wang, J.; Yue, L.; Li, Z.; Zhang, J.; Tian, H.; Willner, I. 2019) also provide interesting annotations and quotes about the possibilities of the origami technique and the design of DNA walkers with motor capacity to start movement, turn and stop, according to molecular interaction patterns. In fact, according to ( Lund, K.; Manzo, AJ; Dabby, N.; Michelotti, N.; Johnson-Buck, A.; Nangreave, J.; Yan, H. 2010 ) these DNA walkers are essentially Molecular robots guided by substrate molecules (precursors) in a set of origami DNA structures (templates). This is confirmed in the following full-text quote from Lund, also corroborated by (Omabegho, T.; Sha, R.; Seeman, NC 2009 | Gu, H.; Chao, J.; Xiao, SJ;Seeman, NC 2010):
“Moving robotics to the level of a single molecule is possible a priori, but it requires facing the limited capacity of individual molecules to store information and complex programs. One strategy to overcome this problem is to use systems that can obtain complex behaviors from the interaction of simple robots with their environment. A first step in this direction was the development of DNA walkers, which have gone from being non-autonomous, to being able to perform brief but directed movements on one-dimensional tracks. In this work we show that random walkers, also called molecular spiders that comprise a streptavidin molecule as an inert -body- and three deoxyribozymes as catalytic -paws-, they exhibit elementary robotic behavior when interacting with a precisely defined environment. Single-molecule microscopy observations confirm that these walkers achieve directional motion by detecting and modifying the tracks of substrate molecules arranged in a two-dimensional DNA origami landscape.” (Lund, K .; Manzo, AJ; Dabby, N .; Michelotti, N .; Johnson-Buck, A .; Nangreave, J .; Yan, H. 2010 )
This can confirm the presence of molecules and pieces with the capacity for self-assembly, their movement, orientation and self-organization, to configure complex electronic devices, according to the patterns and templates of synthetic DNA, that are closer in a solution such as of the vaccine, as suggested by the observation in video 1.
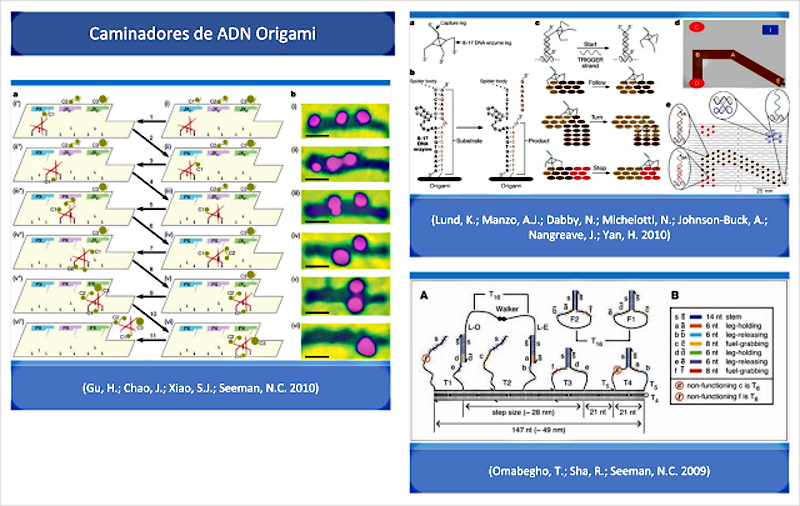
Continuing with the analysis of (Wang, J.; Yue, L.; Li, Z.; Zhang, J.; Tian, H.; Willner, I. 2019), it is added that “The functionalization of edges of the mosaics of origami (from DNA templates), was applied to design programmed multi-component origami structures and, in particular, to develop interchangeable origami dimers .” In other words, DNA templates can be defined in such a way that they are made up of specific pieces (particles, proteins, quantum dots, etc.) according to a predetermined program or pattern.
However, technology of origami DNA may cover additional areas, as stated experiences the state of the Wang and his team “s and manufactured ingenious systems of origami 3D. For example, demonstrated self – assembly of a box of origami, the staggered assembly of gigadalton -scale programmable DNA structures, and the light-driven movement of 3D origami packages to produce reversible chiroptic functions. Different applications of origami nanostructures were suggested, including programmed catalysis, controlled release of drugs, logic gate operations and detection“. Among the applications mentioned, it is worth highlighting the logic gate and detection operations, typical of the QCA (Quantum Cell Automata) circuit design already commented on in the identification of nanorouters among the patterns observed in vaccines. This is one more proof that DNA origami methodology is valid for developing electronic devices based on quantum dots, given the ability to control the orderly construction of cables and circuits.
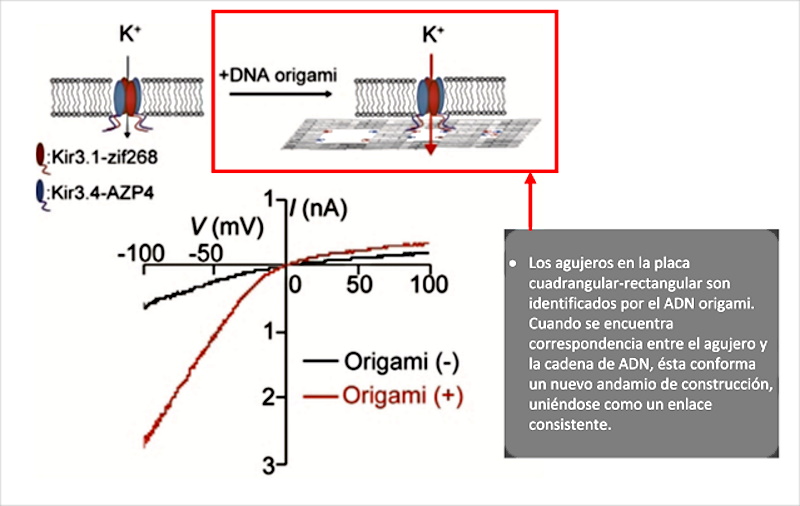
After completing the review of the preambles of the article by (Wang, J.; Yue, L.; Li, Z.; Zhang, J.; Tian, H.; Willner, I. 2019), the scientific discourse focuses on the object of the cavities or holes in the “origami rafts”, which in the shot are shown as quadrangular structures with a dot inscribed within their area. As indicated “Most of these functional origami structures involved, bottom-up modification of origami rafts, edge modification of origami tiles, or folding of the tiles into tubes. However, functionalization of origami structures with nanocavities (holes or barrels) that could act as containment or channels for guided chemical transformations can be considered. To date, such cavities have been fabricated within the passive assembly of origami tiles and these cavities have been used for site-specific coupling of antibodies, reconstitution of membrane proteins, and functionalization of solid-state pores for selective transport. What’s more, DNA structures (not origami) have been introduced into the membranes and these acted as channels for the stimulated potential transport of cargo species across the membranes. On the contrary, the present study introduces the concept of active manufacturing of nanoholes in origami tiles. We report on the active DNAzyme-guided formation of nanoholes in origami scaffolds and the molecular mechanical unlocking of nanoholes by lifting the covered window domains. By applying two different DNAzymes, the programmed and activated fabrication of nanoholes in origami structures is demonstrated. In addition, we use the cavities in the different origami scaffolds as confined nanoenvironments for selective and specific catalysis. What’s more, we highlight a design for the reversible mechanical opening and closing by light of the nanoholes, and the switchable catalysis in the nanocavities. We report on the active DNAzyme-guided formation of nanoholes in origami scaffolds and the molecular mechanical unlocking of nanoholes by lifting the covered window domains. By applying two different DNAzymes, the programmed and activated fabrication of nanoholes in origami structures is demonstrated. In addition, we use the cavities in the different origami scaffolds as confined nanoenvironments for selective and specific catalysis. What’s more, We highlight a design for the reversible mechanical opening and closing by light of the nanoholes, and the switchable catalysis in the nanocavities. We report on the active DNAzyme-guided formation of nanoholes in origami scaffolds and the molecular mechanical unlocking of nanoholes by lifting the covered window domains. By applying two different DNAzymes, the programmed and activated fabrication of nanoholes in origami structures is demonstrated. In addition, we use the cavities in the different origami scaffolds as confined nanoenvironments for selective and specific catalysis. What’s more, We highlight a design for the reversible mechanical opening and closing by light of the nanoholes, and the switchable catalysis in the nanocavities we use the cavities in the different origami scaffolds as confined nanoenvironments for selective and specific catalysis. What’s more, we highlight a design for the reversible mechanical opening and closing by light of the nanoholes, and the switchable catalysis in the nanocavities we use the cavities in the different origami scaffolds as confined nanoenvironments for selective and specific catalysis. What’s more, We highlight a design for the reversible mechanical opening and closing by light of the nanoholes, and the switchable catalysis in the nanocavities.” In this explanation, which leaves no doubt as to the intentionality of the origami technique, there is a fundamental detail that must be seriously considered. It is the ability of cavities in DNA origami structures to trap, immobilize and couple antibodies ( Ouyang, X .; De-Stefano, M.; Krissanaprasit, A.; Bank-Kodal, AL; Bech-Rosen, C.; Liu, T.; Gothelf, KV 2017), which was originally intended to be used for serological studies, but applied to the construction of intracorporeal micro / nano scale electronic devices, could achieve the objective of avoiding phagocytization and immobilization of self-shaped structures. It is also revealed that these holes play a very important role in the interaction with other origami DNA sequences, which can fit together (as if it were a Lego piece) to add new construction scaffolding, as explained ( Kurokawa, T. ; Kiyonaka, S .; Nakata, E .; Endo, M .; Koyama, S .; Mori, E .; Mori, Y. 2018 ) in figure 7.
Bibliography
- Abbasi, QH; Yang, K .; Chopra, N .; Jornet, JM; Abuali, NA; Qaraqe, KA; Alomainy, A. (2016). Nano-communication for biomedical applications: A review on the state-of-the-art from physical layers to novel networking concepts. IEEE Access, 4, pp. 3920-3935. https://doi.org/10.1109/ACCESS.2016.2593582
- Burns, JR; Seifert, A .; Fertig, N .; Howorka, S. (2016). A biomimetic DNA-based channel for the ligand-controlled transport of charged molecular cargo across a biological membrane. Nature nanotechnology, 11 (2), pp. 152-156. https://doi.org/10.1038/nnano.2015.279
- Burns, MA; Mastrangelo, CH; Sammarco, TS; Man, FP; Webster, JR; Johnsons, BN; Burke, DT (1996). Microfabricated structures for integrated DNA analysis = Microfabricated structures for integrated DNA analysis. Proceedings of the National Academy of Sciences, 93 (11), pp. 5556-5561. https://doi.org/10.1073/pnas.93.11.5556
- Campra, P. (2021a). Observations of possible microbiotics in COVID RNAm Version 1. vaccines http://dx.doi.org/10.13140/RG.2.2.13875.55840
- Campra, P. (2021b). Detection of graphene in COVID19 vaccines by Micro-RAMAN spectroscopy. https://www.researchgate.net/publication/355684360_Deteccion_de_grafeno_en_vacunas_COVID19_por_espectroscopia_Micro-RAMAN
- Campra, P. (2021c). MICROSTRUCTURES IN COVID VACCINES: Inorganic crystals or Wireless Nanosensors Network? https://www.researchgate.net/publication/356507702_MICROSTRUCTURES_IN_COVID_VACCINES_inorganic_crystals_or_Wireless_Nanosensors_Network
- Catania, V .; Mineo, A .; Monteleone, S .; Patti, D. (2014). Distributed topology discovery in self-assembled nano network-on-chip. Computers & Electrical Engineering, 40 (8), pp. 292-306. https://doi.org/10.1016/j.compeleceng.2014.09.003
- Chen, Y .; Pepin, A. (2001). Nanofabrication: conventional and non-conventional methods = Nanofabrication: Conventional and nonconventional methods. Electrophoresis, 22 (2), pp. 187-207. https://doi.org/10.1002/1522-2683(200101)22:2%3C187::AID-ELPS187%3E3.0.CO;2-0
- Delgado, R. (2021). Direct from the contents of a Pfizer vaccine under the microscope [Special program]. Fifth column. https://www.twitch.tv/videos/1245191848
- Endo, M .; Sugiyama, H. (2014). Single-molecule imaging of dynamic motions of biomolecules in DNA origami nanostructures using high-speed atomic force microscopy. Accounts of chemical research, 47 (6), pp. 1645-1653. https://doi.org/10.1021/ar400299m
- Esener, SC; Hartmann, DM; Heller, MJ; Cable, JM (1998). DNA-assisted microassembly: a heterogeneous integration technology for optoelectronics = DNA-assisted microassembly: a hetrogeneous integration technology for optoelectronics. In: Heterogeneous Integration: Systems on a Chip: A Critical Review (Vol. 10292, p. 1029208). International Society for Optics and Photonics. https://doi.org/10.1117/12.300616
- Gabrys, PA; Seo, SE; Wang, MX; Oh, E .; Macfarlane, RJ; Mirkin, CA (2018). Lattice mismatch in crystalline nanoparticle thin films = Lattice mismatch in crystalline nanoparticle thin films. Nano letters, 18 (1), pp. 579-585. https://doi.org/10.1021/acs.nanolett.7b04737
- Galal, A .; Hesselbach, X. (2018). Nano-networks communication architecture: Modeling and functions = Nano-networks communication architecture: Modeling and functions. Nano Communication Networks, 17, pp. 45-62. https://doi.org/10.1016/j.nancom.2018.07.001
- Galal, A .; Hesselbach, X. (2020). Probability-based path discovery protocol for electromagnetic nano-networks. Computer Networks, 174, 107246. https://doi.org/10.1016/j.comnet.2020.107246
- Gu, H .; Chao, J .; Xiao, SJ; Seeman, NC (2010). A proximity-based programmable DNA nanoscale assembly line. Nature, 465 (7295), pp. 202-205. https://doi.org/10.1038/nature09026
- Hong, F .; Zhang, F .; Liu, Y .; Yan, H. (2017). DNA origami: scaffolds for creating higher order structures. Chemical reviews, 117 (20), pp. 12584-12640. https://doi.org/10.1021/acs.chemrev.6b00825
- Keren, K .; Berman, RS; Buchstab, E .; Sivan, U .; Braun, E. (2003). DNA-Templated Carbon Nanotube Field-Effect Transistor = DNA template carbon nanotube field effect transistor. Science, 302 (5649), pp. 1380-1382. https://doi.org/10.1126/science.1091022 | https://sci-hub.yncjkj.com/10.1126/science.1091022
- Krahne, R .; Yacoby, A .; Shtrikman, H .; Bar-Joseph, I .; Dadosh, T .; Sperling, J. (2002). Fabrication of nanoscale gaps in integrated circuits = Fabrication of nanoscale gaps in integrated circuits. Applied physics letters, 81 (4), pp. 730-732. https://doi.org/10.1063/1.1495080
- Kumar, P. (2010). Directed self-assembly: expectations and achievements. Nanoscale research letters, 5 (9), pp. 1367-1376. https://doi.org/10.1007/s11671-010-9696-9 | https://sci-hub.yncjkj.com/10.1007/s11671-010-9696-9
- Kurokawa, T .; Kiyonaka, S .; Nakata, E .; Endo, M .; Koyama, S .; Mori, E .; Mori, Y. (2018). DNA Origami scaffolds as templates for functional tetrameric Kir3 K + channels. Angewandte Chemie International Edition, 57 (10), pp. 2586-2591. https://doi.org/10.1002/anie.201709982
- Lewis, DJ; Zornberg, LZ; Carter, DJ; Macfarlane, RJ (2020). Monocrystalline Winterbottom constructions of nanoparticle superlattices = Single-crystal Winterbottom constructions of nanoparticle superlattices. Nature materials, 19 (7), pp. 719-724. https://doi.org/10.1038/s41563-020-0643-6
- Liu, J .; Lu, Y. (2003). A colorimetric lead biosensor using DNAzyme-directed assembly of gold nanoparticles = A colorimetric lead biosensor using DNAzyme-directed assembly of gold nanoparticles. Journal of the American Chemical Society, 125 (22), pp. 6642-6643. https://doi.org/10.1021/ja034775u
- Liu, J .; Lu, Y. (2006). Fast colorimetric sensing of adenosine and cocaine based on a general sensor design involving aptamers and nanoparticles. Angewandte Chemie, 118 (1), pp. 96-100. https://doi.org/10.1002/ange.200502589
- Liu, J .; Wei, J .; Yang, Z. (2021). Building ordered nanoparticle assemblies inspired by atomic epitaxy. Physical Chemistry Chemical Physics, 23 (36), pp. 20028-20037. https://doi.org/10.1039/D1CP02373J | https://sci-hub.yncjkj.com/10.1039/D1CP02373J
- Lund, K .; Manzo, AJ; Dabby, N .; Michelotti, N .; Johnson-Buck, A .; Nangreave, J .; Yan, H. (2010). Molecular robots guided by prescriptive landscapes = Molecular robots guided by prescriptive landscapes. Nature, 465 (7295), pp. 206-210. https://doi.org/10.1038/nature09012
- McMillan, RA; Paavola, CD; Howard, J .; Chan, SL; Zaluzec, NJ; Trent, JD (2002). Ordered nanoparticle arrays formed on engineered chaperonin protein templates. Nature materials, 1 (4), pp. 247-252. https://doi.org/10.1038/nmat775
- Nykypanchuk, D .; Maye, MM; Van-Der-Lelie, D .; Gang, O. (2008). DNA-guided crystallization of colloidal nanoparticles = DNA-guided crystallization of colloidal nanoparticles. Nature, 451 (7178), pp. 549-552. https://doi.org/10.1038/nature06560
- Omabegho, T .; Sha, R .; Seeman, NC (2009). A bipedal DNA Brownian motor with coordinated legs = A bipedal DNA Brownian motor with coordinated legs. Science, 324 (5923), pp. 67-71. https://doi.org/10.1126/science.1170336
- Oukhatar, A .; Bakhouya, M .; El Ouadghiri, D. (2021). Electromagnetic-Based Wireless Nano-Sensors Network: Architectures and Applications. J. Commun., 16 (1), 8. https://www.researchgate.net/profile/Driss-El-Ouadghiri/publication/348355577…/Electromagnetic-Based-Wireless-Nano-Sensors-Network-Architectures -and-Applications.pdf
- Ouyang, X .; De-Stefano, M .; Krissanaprasit, A .; Bank-Kodal, AL; Bech-Rosen, C .; Liu, T .; Gothelf, KV (2017). Docking of antibodies into the cavities of DNA origami structures. Angewandte Chemie, 129 (46), pp. 14615-14619. https://doi.org/10.1002/ange.201706765
- Rothemund, PW (2006). Folding DNA to create nanoscale shapes and patterns. Nature, 440 (7082), pp. 297-302. https://doi.org/10.1038/nature04586
- Sarlangue, G .; Devilleger, J .; Trillaud, P .; Fouchet, S .; Taillasson, L .; Catteau, G. (2021). Objectification of the existence of discoverable MAC addresses in the Bluetooth frequency range after an inoculation of COVID antigen therapy and the COVID detection PCR test. https://ln5.sync.com/dl/195df4a10/5ab9apq6-q5vgawam-vgr3ktt9-7zr985rh/view/default/2451906512011
- Seifert, A .; Göpfrich, K .; Burns, JR; Fertig, N .; Keyser, UF; Howorka, S. (2015). Bilayer DNA nanopores with voltage-switching between open and closed state = Bilayer-spanning DNA nanopores with voltage-switching between open and closed state. ACS nano, 9 (2), pp. 1117-1126. https://doi.org/10.1021/nn5039433
- Shen, J .; Sun, W .; Liu, D .; Schaus, T .; Yin, P. (2021). Three-dimensional nanolithography guided by DNA modular epitaxy = Three-dimensional nanolithography guided by DNA modular epitaxy. Nature Materials, 20 (5), pp. 683-690. https://doi.org/10.1038/s41563-021-00930-7
- Taton, TA; Mirkin, CA; Letsinger, RL (2000). Scanometric DNA array detection with nanoparticle probes = Scanometric DNA array detection with nanoparticle probes. Science, 289 (5485), pp. 1757-1760. https://doi.org/10.1126/science.289.5485.1757
- Wang, J .; Yue, L .; Li, Z .; Zhang, J .; Tian, H .; Willner, I. (2019). Active generation of nanoholes in DNA origami scaffolds for programmed catalysis in nanocavities = Active generation of nanoholes in DNA origami scaffolds for programmed catalysis in nanocavities. Nature communications, 10 (1), pp. 1-10. https://doi.org/10.1038/s41467-019-12933-9 | https://www.researchgate.net/publication/336927899_Active_generation_of_nanoholes_in_DNA_origami_scaffolds_for_programmed_catalysis_in_nanocavities
- Wang, MX; Seo, SE; Gabrys, PA; Fleischman, D .; Lee, B .; Kim, Y .; Mirkin, CA (2017). Epitaxy: Programmable atom equivalents versus atoms = Epitaxy: Programmable atom equivalents versus atoms. ACS nano, 11 (1), pp. 180-185. https://doi.org/10.1021/acsnano.6b06584
- Yang, J .; Ma, M .; Li, L .; Zhang, Y .; Huang, W .; Dong, X. (2014). Graphene nanomesh: new versatile materials = Graphene nanomesh: new versatile materials. Nanoscale, 6 (22), pp. 13301-13313. https://doi.org/10.1039/C4NR04584J | https://sci-hub.yncjkj.com/10.1039/c4nr04584j
- Zhang, R .; Yang, K .; Abbasi, QH; Qaraqe, KA; Alomainy, A. (2017). Analytical characterization of the terahertz in-vivo nano-network in the presence of interference based on TS-OOK communication scheme. IEEE Access, 5, pp. 10172-10181. https://doi.org/10.1109/ACCESS.2017.2713459
Connect with Mik Andersen at Corona2Inspect
Click here to see related articles featuring the work of La Quinta Columna
See related by Mik Andersen: